Proteins are complex molecules that play crucial roles in various biological processes. In order for proteins to function effectively, they must have the correct three-dimensional structure. Traditional methods of comparing protein structures have focused solely on the position of atoms, neglecting important chemical information. However, a new groundbreaking technique called LoCoHD (Local Composition Hellinger Distance) developed by the HUN-REN-ELTE Protein Modeling Research Group takes into account both the position of atoms and their chemical properties. This method has the potential to revolutionize the field of protein structure analysis.
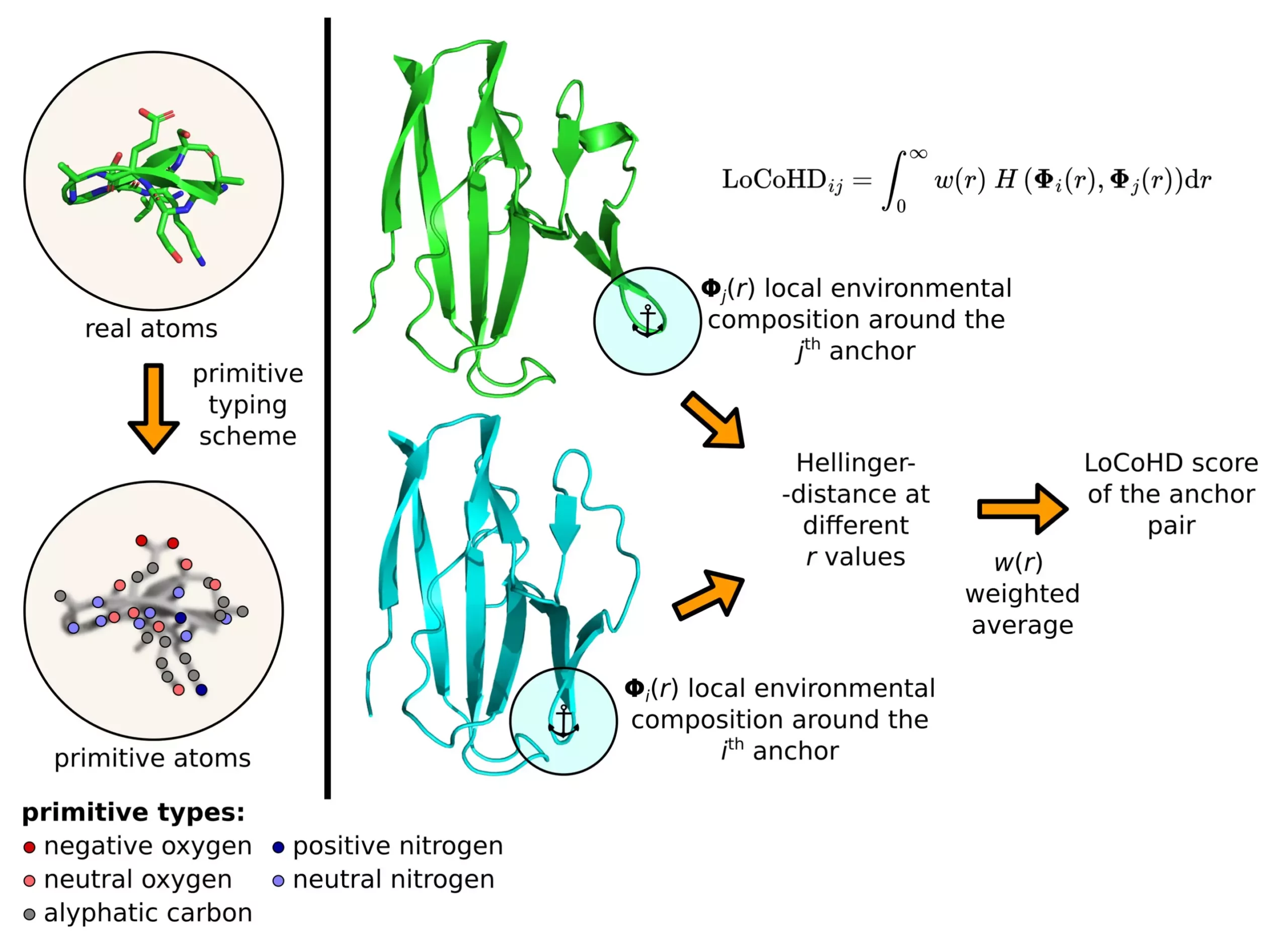
The LoCoHD algorithm, developed by Zsolt Fazekas, is a unique approach to comparing local environments around amino acids in proteins based on their chemical nature. By considering factors such as elemental composition, charge, and hydrophobicity, the algorithm assigns a value between 0 and 1 to indicate the similarity or dissimilarity of protein structures. A value close to 0 suggests high similarity, while a value close to 1 indicates significant differences in properties. This numerical metric provides valuable insights into the structural characteristics of proteins.
The LoCoHD algorithm employs a multi-step protocol to generate a numerical representation of structural differences between proteins. In the initial step, real atoms in the protein are converted into primitive atoms, each representing a specific chemical nature (e.g., positively charged nitrogen, negatively charged oxygen, etc.). The second step involves selecting anchor atoms as reference points for comparison. By analyzing dissimilarity measures between pairs of anchor atoms, the algorithm generates a comprehensive descriptor of the entire protein structure. This approach enables researchers to gain a deeper understanding of protein conformation and function.
The LoCoHD method has broad applications in protein research, including its use in the biannual CASP competitions, where participants predict protein structures based on unpublished data. By incorporating chemical information into structure comparison techniques, the LoCoHD algorithm offers a novel perspective on protein analysis. In a recent study focusing on the SARS-CoV-2 virus protein ORF8, researchers demonstrated the effectiveness of LoCoHD in identifying critical differences in amino acid environments between predicted and experimental structures. This innovative approach has the potential to enhance our understanding of disease-causing pathogens and facilitate the development of targeted therapies.
Beyond static structure analysis, the LoCoHD method has proven effective in studying the internal motion of proteins. Using simulations to simulate molecular movements, researchers identified key amino acids undergoing significant chemical changes during protein motion. For instance, in the study of the podocin protein, essential for kidney function, the algorithm pinpointed amino acids critical for structural stability and function. Similarly, in the investigation of the HIV-1 capsid protein, LoCoHD identified an amino acid crucial for viral envelope formation. These findings not only expand our knowledge of protein dynamics but also hold promise for the development of targeted therapies against various diseases.
The development of the LoCoHD algorithm represents a significant advancement in protein structure analysis. By integrating chemical information into structural comparisons, this method offers a comprehensive view of protein characteristics and dynamics. From predicting protein structures to studying molecular movements, the applications of LoCoHD are vast and promising. As researchers continue to explore the potential of this innovative approach, we can expect further insights into the complex world of proteins and their roles in health and disease.

Leave a Reply