Protein synthesis is a fundamental process in cells that is essential for their survival. Ribosomes, which act as the cell’s “protein factories,” play a crucial role in this process. They are large molecules found in all living organisms. Ribosomes read the blueprint for a specific protein on a messenger molecule called messenger RNA (mRNA) and convert this information into a new protein. Ribosomes consist of two subunits: the small subunit, responsible for reading and checking the mRNA for errors, and the large subunit, responsible for the polymerization of amino acids to form proteins.
While the assembly of ribosomes has been increasingly understood in recent years, the process of ribosomal degradation remains poorly understood. In stress situations, such as a lack of food or at the end of their growth cycle, cells reduce their metabolism to survive longer. This state is known as the stationary phase. During this phase, energy-intensive protein synthesis is reduced, and some ribosomes are degraded to release the energy invested in them, ensuring cell survival.
A research team from the Department of Chemistry at the Universität Hamburg has made a significant breakthrough in understanding the molecular mechanism of ribosomal degradation. The team focused on the enzyme RNase R, known to be involved in the degradation process of ribosomes during stress situations.
The researchers studied Bacillus subtilis, a rod-shaped soil bacterium, to gain insight into the processes occurring during the transition to the stationary phase. They specifically chose cells that were still growing rather than in the stationary phase to explore the early stages of ribosomal degradation.
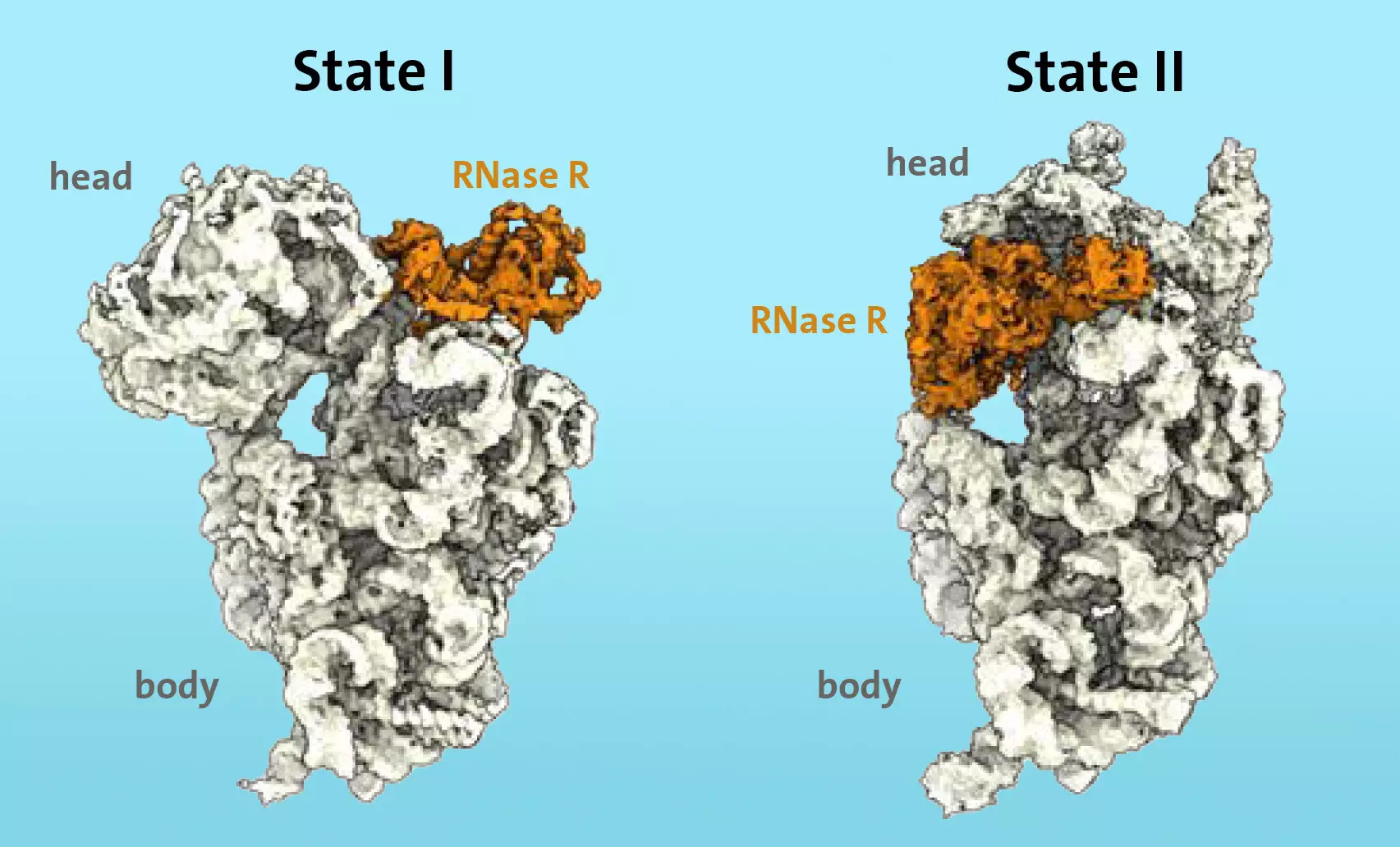
Using cryo-electron microscopy, the researchers discovered that the enzyme RNase R binds to the small 30S subunit of the ribosome. The “S” in 30S stands for “Svedberg units,” which refers to the mass of the ribosomal subunit. The researchers observed that RNase R does not randomly cut the 30S subunit but instead attaches itself to a specific area called the “neck” and then detaches the “head,” the upper area of the subunit, in two consecutive stages.
During the first stage, the enzyme encounters an obstacle at the “neck” and destabilizes the area, making it more flexible. In the second stage, the “head” is turned, removing the obstacle and allowing the enzyme to continue the degradation process unhindered.
The research team’s in-vitro degradation experiments showed that the “head” switch serves as a significant kinetic barrier for RNase R. Additionally, they demonstrated that the enzyme alone is sufficient to complete the degradation process of the 30S subunit.
This study provides crucial structural insights into the degradation mechanism of ribosomes, shedding light on a process that was previously poorly understood. Understanding how ribosomes are degraded is essential for comprehending the cellular response to stress and the survival mechanisms employed by cells.
The findings of this study open up new avenues for further research into ribosomal degradation. Investigating other enzymes involved in the process, as well as exploring how different stress conditions impact ribosomal degradation, can provide a more comprehensive understanding of cellular survival mechanisms.
The research team from the Universität Hamburg has made significant strides in elucidating the dynamic mechanism used by the enzyme RNase R to degrade the ribosomal 30S subunit. By studying a rod-shaped soil bacterium and using cryo-electron microscopy, the team observed how RNase R binds to the 30S subunit and initiates the degradation process by destabilizing the “neck” area. These findings contribute to our understanding of cellular responses to stress and the survival strategies employed by cells. Further research in this field can help uncover additional details about ribosomal degradation, ultimately advancing our knowledge of essential cellular processes.

Leave a Reply