Fungal infections pose a significant challenge to human health, with over 300,000 people affected by Aspergillosis, a life-threatening disease caused by the fungal pathogen Aspergillus fumigatus, each year. In a recent study published in Chemical Science, investigators at the University of Kansas have made a significant breakthrough in understanding the genes responsible for producing sartorypyrones, a chemical produced by A. fumigatus and its fungal cousins. These findings could potentially lead to the development of more effective antifungal drugs and better therapeutic interventions.
The Challenge of Fungal Infections
Fungal infections, especially those caused by Aspergillus fumigatus, are particularly problematic for individuals with compromised immune systems, such as cancer patients undergoing treatment or those living in sub-Saharan Africa without access to proper medication. These infections have gained increased attention in both scientific and media reports, highlighting the urgent need for better preventive measures and treatments.
Exploring Fungal Secondary Metabolites
Secondary metabolites produced by fungi, including A. fumigatus, have long been of interest for their potential medicinal properties. However, studying these metabolites in the laboratory setting presents numerous challenges. These compounds are biologically active and offer selective advantages to the fungi, such as protection against UV radiation and competition inhibition. While some secondary metabolites have beneficial bioactivities, others contribute to the pathogenic effects of the fungus, including immune system suppression.
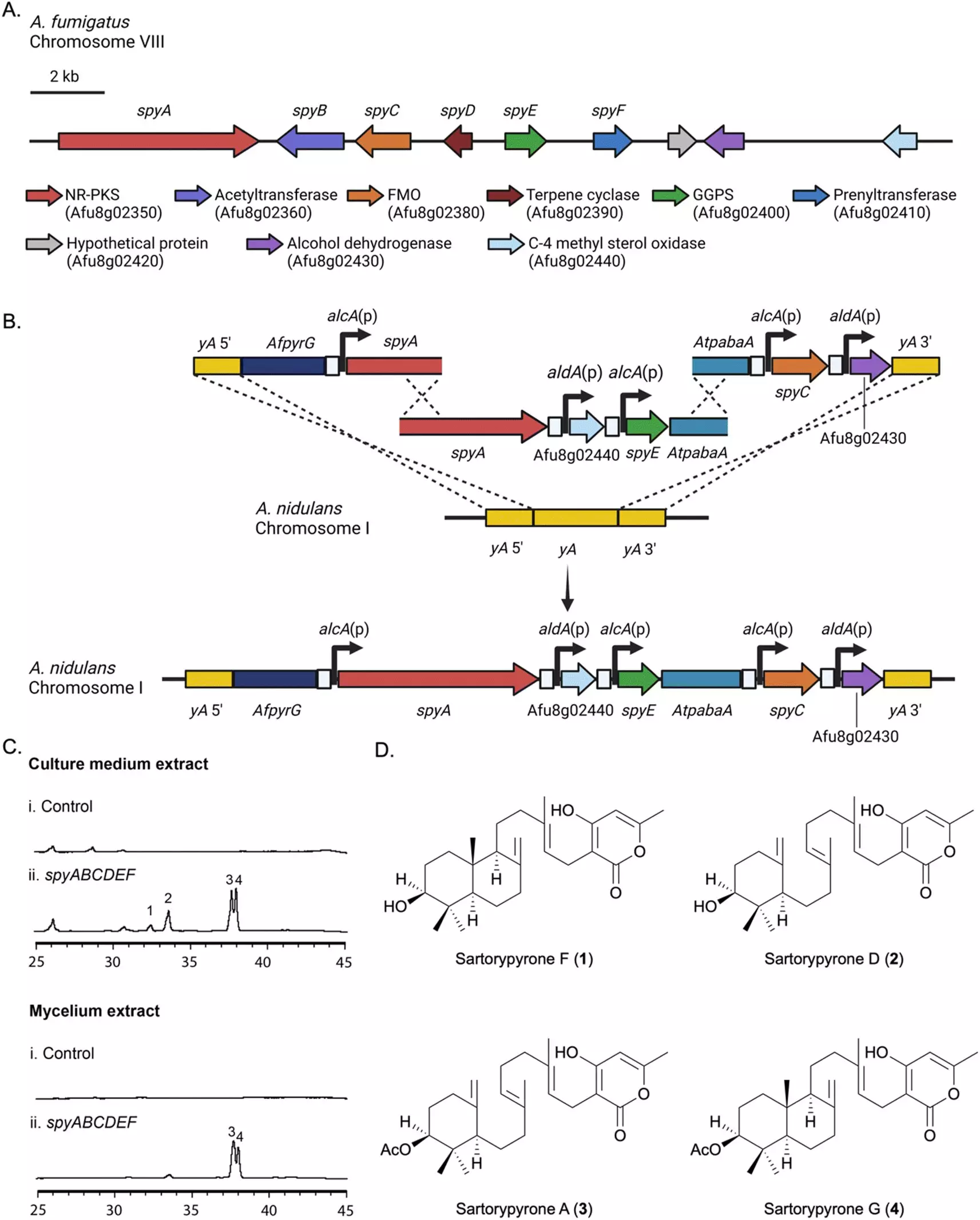
To better understand the genes responsible for the production of secondary metabolites in A. fumigatus, the research team at the University of Kansas transferred a biosynthetic gene cluster (BGC) to a related strain of Aspergillus called A. nidulans. This technique, known as “heterologous expression,” allows the team to observe the compounds produced in the lab. In this particular study, a gene cluster responsible for sartorypyrone synthesis, named the “spy BGC,” was analyzed using advanced techniques such as high-resolution electrospray ionization mass spectrometry, nuclear magnetic resonance, and microcrystal electron diffraction (MicroED).
Through their analysis, the team was able to identify 12 chemical products from the spy BGC, including seven compounds that had not been isolated previously. This discovery provides valuable insights into the biosynthetic pathway of sartorypyrones and offers a straightforward approach to dissecting the overall biosynthetic pathway. The researchers propose that this methodology can be applied to other gene clusters in A. fumigatus, opening up new possibilities for the understanding of fungal pathogens and the development of targeted therapies.
The findings of this study have the potential to lead to new therapies for fungal infections and eco-friendly industrial applications. Understanding the secondary metabolite gene clusters in A. fumigatus can help researchers uncover the mechanisms of infection and devise strategies to limit its impact. Furthermore, this knowledge can be leveraged to develop advanced antifungal medications. However, the economic realities of manufacturing antifungal drugs may present challenges, as the current pharmaceutical industry prioritizes highly profitable long-term medications over short-term solutions.
The University of Kansas researchers’ breakthrough in deciphering the gene cluster responsible for sartorypyrone production in Aspergillus fumigatus represents a significant step forward in understanding fungal infections. By exploring secondary metabolites and their biosynthetic pathways, scientists can develop new therapies and interventions to combat diseases caused by pathogenic fungi. While challenges exist in translating these discoveries into practical medications, continued research in this field holds promise for improving the lives of those affected by fungal infections.

Leave a Reply