In a groundbreaking development, a research team has introduced a new technique that could potentially transform the way scientists identify novel targets for cancer therapies. The new method, known as EndoVIA, has the potential to provide unprecedented insights into the editing process of RNA in living cells. This breakthrough has the potential to unravel the mystery behind various illnesses, particularly cancer.
The language of RNA plays a critical role in translating our genetic code into proteins within our cells. However, these RNA messages are constantly in need of editing to prevent diseases such as cancer. While the editing process is essential for maintaining good health, it has always been challenging to observe it in intricate detail.
Led by graduate student Alex Quillin, a team of WashU chemists has successfully created a test called EndoVIA that enables researchers to precisely track individual edits within cells. This innovative approach has the potential to revolutionize our understanding of how diseases originate and progress. The groundbreaking findings of this research were recently published in the prestigious journal ACS Central Science.
RNA editing is of utmost importance for the proper functioning of cells, as any abnormalities in this process can lead to various diseases. Despite the significance of RNA editing, little is known about the precise timing, location, and regulation of these edits within cells. The most common type of RNA editing involves the conversion of adenosine molecules into inosine through a specific enzyme. This natural process is believed to aid in distinguishing between normal RNA and foreign RNA, triggering an immune response when necessary.
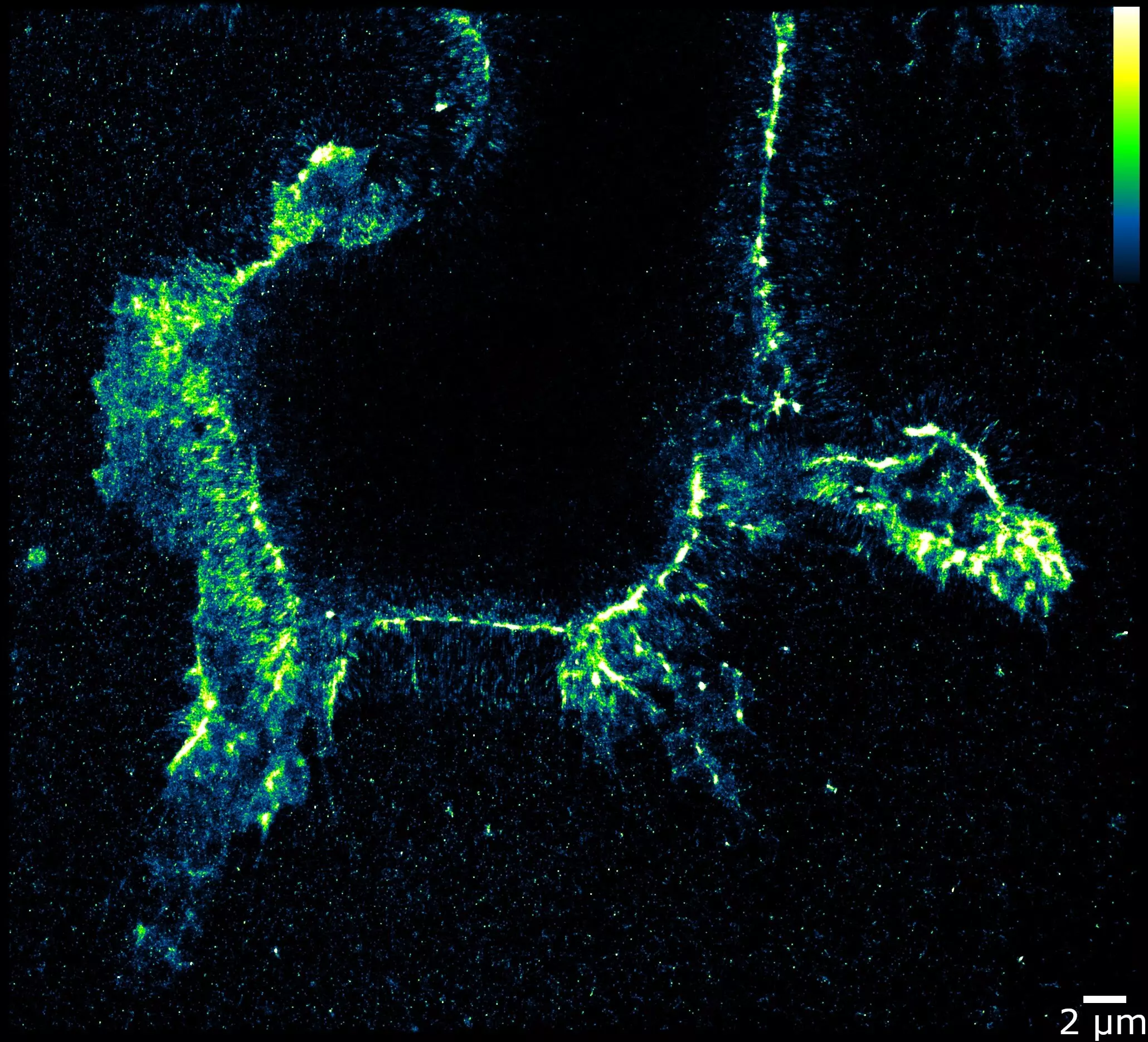
Traditionally, tracking RNA edits required isolating RNA from cells, combining the molecules, and subjecting the sample to sequencing analysis. While this method allowed for the quantification of edits, it lacked the ability to pinpoint the exact location of the edits within the cell. The WashU research team has repurposed an enzyme that targets edited RNAs, enabling researchers to visualize and track these critical edits. By replacing magnesium with calcium, the enzyme binds to the edited RNA elements in cells, which can then be tagged with fluorescent antibodies for microscopic observation.
The innovative EndoVIA technique developed by the WashU team has the potential to revolutionize cancer therapy by aiding scientists in identifying novel targets for treatment. This is particularly crucial as certain types of cancers have been found to be associated with abnormalities in RNA editing. For example, breast cancer cells often exhibit excessive editing, while kidney cancer cells may have insufficient editing. In a recent study, researchers utilized EndoVIA to observe differences in RNA editing patterns between healthy and cancerous kidney cells, shedding light on potential targets for future investigations.
The groundbreaking technique developed by the WashU research team holds immense promise in advancing our understanding of RNA editing and its implications for diseases such as cancer. By enabling researchers to visualize individual RNA molecules within cells with unprecedented clarity, EndoVIA opens up new avenues for studying the biological causes and effects of RNA editing abnormalities. This innovative approach has the potential to transform cancer therapy and pave the way for novel treatment strategies based on a deeper understanding of RNA editing processes.

Leave a Reply