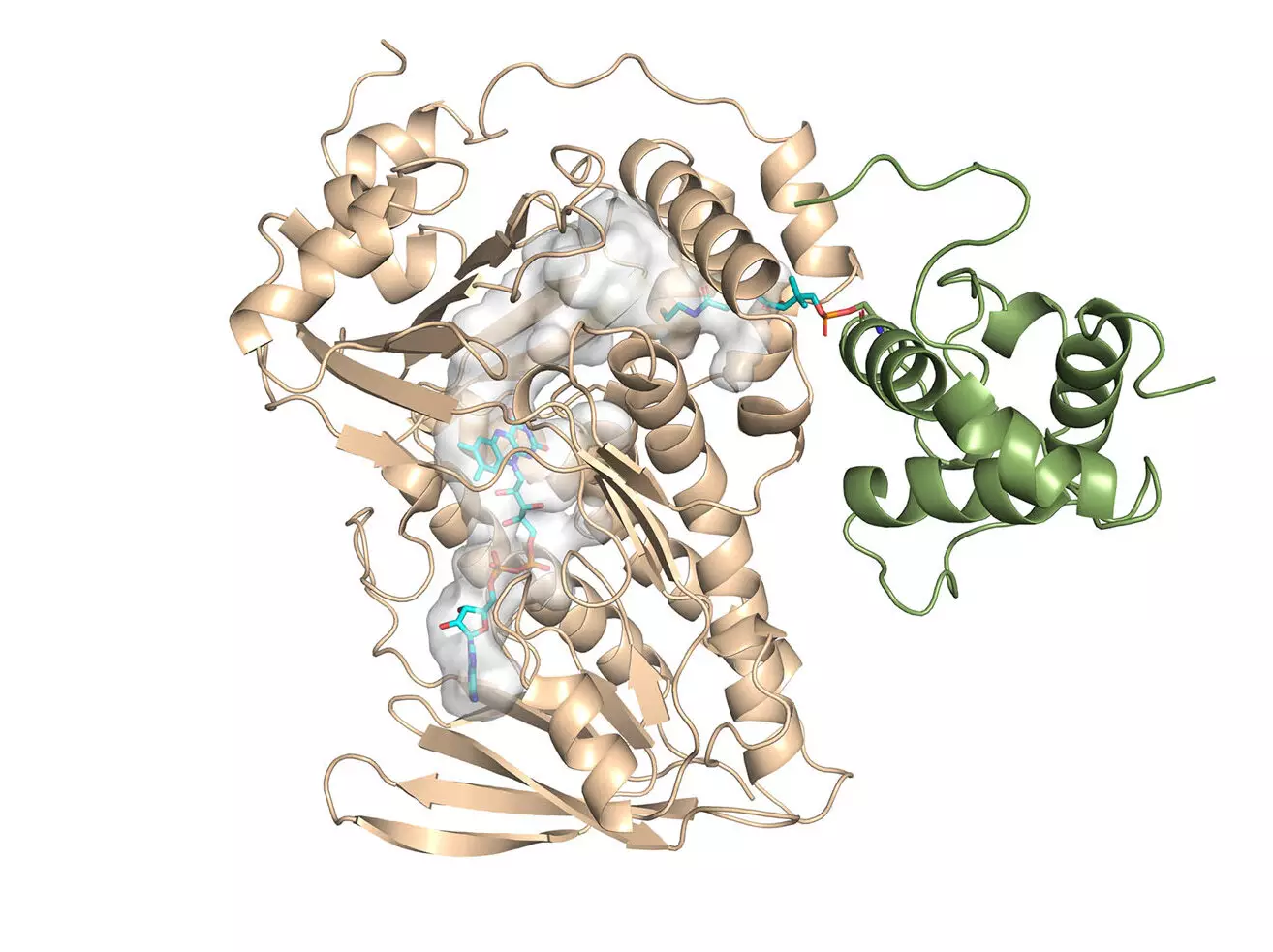
In a groundbreaking study, chemists have successfully determined the crystal structure of a key component of the monensin enzyme, offering valuable insights into its reaction activity and paving the way for the development of safer and more effective antibiotics. Led by Professor Chu-Young Kim from the University of Illinois Urbana-Champaign (UIUC), the research team unlocked the mechanism of this important enzyme by solving the crystal structure for MonCI, a critical enzyme found in soil bacteria that naturally synthesizes monensin. Collaborating with the team, Associate Professor Lela Vukovic from The University of Texas at El Paso (UTEP) conducted computational studies on the monensin research, which were vital in unraveling its molecular secrets.
Crystal Structure Discovery
The main finding of the study was the groundbreaking discovery of the first-ever crystal structure for this family of enzymes. This accomplishment marks a significant milestone in the field of biochemistry, providing scientists with a deeper understanding of the enzyme’s functions and potential applications. The crystal structure for MonCI not only sheds light on its three crucial epoxidation reactions but also offers valuable insights into how the bacterium can be engineered to produce new antibiotics. The ability to manipulate the enzyme’s structure opens up exciting possibilities for future antibiotic design and development.
To further investigate the enzyme’s mechanisms, Professor Kim and Dr. Vukovic employed computational modeling techniques. Due to the challenges of obtaining the crystal structure of the enzyme with the substrate inside in its active mode experimentally, the team turned to simulations. By modeling the enzyme and substrate in stable positions, they were able to gain insights into the reaction sequence that produces monensin. This computational approach proved indispensable in characterizing the complex sequential reactions that are crucial for antibiotic production. Driven by the power of supercomputers, their research deepened our understanding of these biological molecules and their potential societal impact.
The computational challenges faced by the researchers stemmed from examining multiple systems and determining the optimal positions for premonensin A and its epoxidated versions to undergo the essential first, second, and third epoxidation reactions. While daunting, these challenges were overcome with the aid of advanced supercomputing technologies. The University of Texas Research Cyberinfrastructure (UTRC) initiative provided Dr. Vukovic with supercomputer allocations on the Lonestar6 system at the Texas Advanced Computing Center (TACC). These allocations were instrumental in supporting the intensive computational studies performed by Dr. Vukovic and her students.
Dr. Vukovic’s postdoctoral research at UIUC under the late Klaus Schulten played a crucial role in this study. Dr. Schulten was renowned for his work in computational biology and was among the pioneers of molecular dynamics simulations. His contribution to the development of the NAMD software, which was utilized in this study, continues to benefit researchers worldwide. Through the optimized use of NAMD on TACC’s supercomputers like Lonertar6 and Stampede2, researchers can delve deep into the intricate details of biological molecules and elucidate complex reaction pathways.
While the crystal structure of MonCI provides a significant breakthrough, it is essential to investigate the other enzymes involved in monensin biosynthesis. With at least 14 different enzymes required for monensin production, understanding their structure and functions will be crucial in developing improved versions of monensin. The ultimate goal is to create safer alternatives that can effectively address the needs of cattle and poultry. Moreover, given that monensin is toxic to horses and dogs, accidental poisoning cases in these animals can be catastrophic. Thus, the development of a non-toxic monensin variant is of paramount importance.
The determination of the crystal structure for MonCI and the insights gained from computational studies represent a significant leap forward in our understanding of the monensin enzyme and its potential applications in antibiotic development. This research not only unveils the intricate details of the enzyme’s reaction mechanisms but also sets the stage for future advancements in the field. By leveraging computational technologies and collaborating across institutions, scientists are harnessing the power of innovation to address societal challenges and pave the way for safer and more effective antibiotics.

Leave a Reply